Introduction
This is the vignette for performing population adjustment methods with count data, in order to compare marginal treatment effects when there are cross-trial differences in effect modifiers and limited patient-level data. We will demonstrate how to apply MAIC, STC, G-computation with ML, G-computation with Bayesian inference and multiple imputation marginalisation. The document structure follow the binary data example vignette which should be referred to for more details.
Example analysis
First, let us load necessary packages.
# install.packages("outstandR",
# repos = c("https://statisticshealtheconomics.r-universe.dev", "https://cloud.r-project.org"))
#
# install.packages("simcovariates",
# repos = c("https://n8thangreen.r-universe.dev", "https://cloud.r-project.org"))
library(boot) # non-parametric bootstrap in MAIC and ML G-computation
library(copula) # simulating BC covariates from Gaussian copula
library(rstanarm) # fit outcome regression, draw outcomes in Bayesian G-computation
library(tidyr)
library(dplyr)
library(MASS)
library(outstandR)
library(simcovariates)Data
We first simulate both the IPD and ALD count data. See the binary
data example vignette for more details on how this is implemented. The
difference with that example is that we change the family
argument in gen_data() to
poisson(link = "log"), corresponding to the count data
case. The gen_data() function is available in the simcovariates
package on GitHub.
N <- 200
allocation <- 2/3 # active treatment vs. placebo allocation ratio (2:1)
b_trt <- log(0.17) # conditional effect of active treatment vs. common comparator
b_X <- -log(0.5) # conditional effect of each prognostic variable
b_EM <- -log(0.67) # conditional interaction effect of each effect modifier
meanX_AC <- c(0.45, 0.45) # mean of normally-distributed covariate in AC trial
meanX_BC <- c(0.6, 0.6) # mean of each normally-distributed covariate in BC
meanX_EM_AC <- c(0.45, 0.45) # mean of normally-distributed EM covariate in AC trial
meanX_EM_BC <- c(0.6, 0.6) # mean of each normally-distributed EM covariate in BC
sdX <- c(0.4, 0.4) # standard deviation of each covariate (same for AC and BC)
sdX_EM <- c(0.4, 0.4) # standard deviation of each EM covariate
corX <- 0.2 # covariate correlation coefficient
b_0 <- -0.6 # baseline intercept coefficient ##TODO: fixed value
covariate_defns_ipd <- list(
PF_cont_1 = list(type = continuous(mean = meanX_AC[1], sd = sdX[1]),
role = "prognostic"),
PF_cont_2 = list(type = continuous(mean = meanX_AC[2], sd = sdX[2]),
role = "prognostic"),
EM_cont_1 = list(type = continuous(mean = meanX_EM_AC[1], sd = sdX_EM[1]),
role = "effect_modifier"),
EM_cont_2 = list(type = continuous(mean = meanX_EM_AC[2], sd = sdX_EM[2]),
role = "effect_modifier")
)
b_prognostic <- c(PF_cont_1 = b_X, PF_cont_2 = b_X)
b_effect_modifier <- c(EM_cont_1 = b_EM, EM_cont_2 = b_EM)
num_normal_covs <- length(covariate_defns_ipd)
cor_matrix <- matrix(corX, num_normal_covs, num_normal_covs)
diag(cor_matrix) <- 1
rownames(cor_matrix) <- c("PF_cont_1", "PF_cont_2", "EM_cont_1", "EM_cont_2")
colnames(cor_matrix) <- c("PF_cont_1", "PF_cont_2", "EM_cont_1", "EM_cont_2")
ipd_trial <- simcovariates::gen_data(
N = N,
b_0 = b_0,
b_trt = b_trt,
covariate_defns = covariate_defns_ipd,
b_prognostic = b_prognostic,
b_effect_modifier = b_effect_modifier,
cor_matrix = cor_matrix,
trt_assignment = list(prob_trt1 = allocation),
family = poisson("log"))
ipd_trial$trt <- factor(ipd_trial$trt, labels = c("C", "A"))Similarly, for the aggregate data but with the additional summarise step (see binary data example vignette for code).
This general format of the data sets are in a ‘long’ style consisting of the following.
ipd_trial: Individual patient data
-
PF_*: Patient measurements prognostic factors -
EM_*: Patient measurements effect modifiers -
trt: Treatment label (factor) -
y: Counts
ald_trial: Aggregate-level data
-
variable: Covariate name. In the case of treatment arm sample size this isNA -
statistic: Summary statistic name from mean, standard deviation or sum -
value: Numerical value of summary statistic -
trt: Treatment label. Because we assume a common covariate distribution between treatment arms this isNA
Our data look like the following.
head(ipd_trial)
#> id PF_cont_1 PF_cont_2 EM_cont_1 EM_cont_2 trt y true_eta
#> 1 1 0.75056436 0.95597583 0.3158969 1.19430733 C 4 0.582883524
#> 2 2 0.83246940 0.11789773 0.5094678 0.08180243 A 1 -1.476422106
#> 3 3 1.13401333 1.15036919 1.2342379 0.74771694 A 3 0.005184912
#> 4 4 0.78518298 0.83080451 -0.1625820 0.35223227 C 1 0.520117172
#> 5 5 -0.38547667 0.87070020 0.7213797 -0.03557032 A 1 -1.760974264
#> 6 6 -0.04851591 -0.03013213 0.6580067 0.05853054 A 0 -2.139514435There are 4 correlated continuous covariates generated per subject,
simulated from a multivariate normal distribution. Treatment
trt takes either new treatment A or standard of
care / status quo C. The ITC is ‘anchored’ via C, the
common treatment.
ald_trial
#> # A tibble: 16 × 4
#> variable statistic value trt
#> <chr> <chr> <dbl> <chr>
#> 1 EM_cont_1 mean 0.616 NA
#> 2 EM_cont_1 sd 0.408 NA
#> 3 EM_cont_2 mean 0.635 NA
#> 4 EM_cont_2 sd 0.354 NA
#> 5 PF_cont_1 mean 0.666 NA
#> 6 PF_cont_1 sd 0.398 NA
#> 7 PF_cont_2 mean 0.663 NA
#> 8 PF_cont_2 sd 0.404 NA
#> 9 y mean 1.07 C
#> 10 y sd 1.01 C
#> 11 y sum 77 C
#> 12 y mean 0.438 B
#> 13 y sd 0.750 B
#> 14 y sum 56 B
#> 15 NA N 72 C
#> 16 NA N 128 BIn this case, we have 4 covariate mean and standard deviation values; and the total, average and sample size for each treatment B and C.
In the following we will implement for MAIC, STC, and G-computation methods to obtain the marginal variance and the marginal treatment effect.
Model fitting in R
The outstandR package has been written to be easy to
use and essential consists of a single function,
outstandR(). This can be used to run all of the different
types of model, which we will call strategies. The first two
arguments of outstandR() are the individual and
aggregate-level data, respectively.
A strategy argument of outstandR takes
functions called strategy_*(), where the wildcard
* is replaced by the name of the particular method
required, e.g. strategy_maic() for MAIC. Each specific
example is provided below.
The formula used in this model, passed as an argument to the strategy function is
lin_form <- as.formula("y ~ PF_cont_1 + PF_cont_2 + trt:EM_cont_1 + trt:EM_cont_2")MAIC
As mentioned above, pass the model specific strategy function to the
main outstandR() function, in this case use
strategy_maic().
outstandR_maic <-
outstandR(ipd_trial, ald_trial,
strategy = strategy_maic(
formula = lin_form,
family = poisson(link = "log")))
#> log rate used
#>
#> log rate usedThe returned object is of class outstandR.
outstandR_maic
#> Object of class 'outstandR'
#> Model: poisson
#> Scale: log_relative_risk
#> Common treatment: C
#> Individual patient data study: AC
#> Aggregate level data study: BC
#> Confidence interval level: 0.95
#>
#> Contrasts:
#>
#> # A tibble: 3 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <dbl> <dbl>
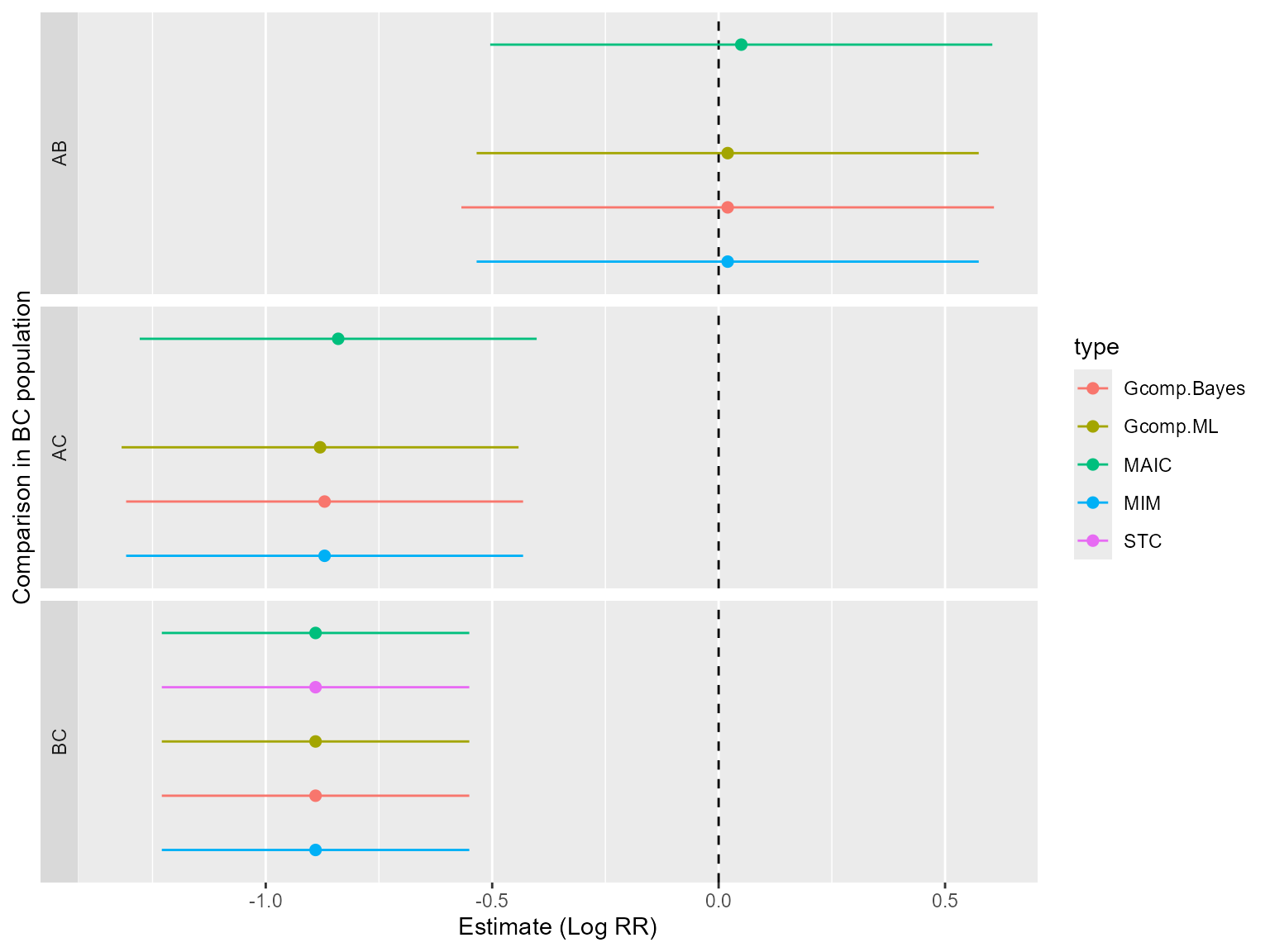
#> 1 AB 0.0524 0.0801 -0.502 0.607
#> 2 AC -0.841 0.0493 -1.28 -0.406
#> 3 BC -0.894 0.0308 -1.24 -0.550
#>
#> Absolute:
#>
#> # A tibble: 2 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <lgl> <lgl>
#> 1 A 0.500 0.00577 NA NA
#> 2 C 1.16 0.0292 NA NASimulated Treatment Comparison (STC)
STC is the conventional outcome regression method. It involves
fitting a regression model of outcome on treatment and covariates to the
IPD. Simply pass the same as formula as before with the
strategy_stc() strategy function.
outstandR_stc <-
outstandR(ipd_trial, ald_trial,
strategy = strategy_stc(
formula = lin_form,
family = poisson(link = "log")))
#> log rate used
#>
#> log rate used
outstandR_stc
#> Object of class 'outstandR'
#> Model: poisson
#> Scale: log_relative_risk
#> Common treatment: C
#> Individual patient data study: AC
#> Aggregate level data study: BC
#> Confidence interval level: 0.95
#>
#> Contrasts:
#>
#> # A tibble: 3 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 AB NaN NA NA NA
#> 2 AC NaN NA NA NA
#> 3 BC -0.894 0.0308 -1.24 -0.550
#>
#> Absolute:
#>
#> # A tibble: 2 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <lgl> <lgl>
#> 1 A NaN NA NA NA
#> 2 C 0.330 NA NA NAParametric G-computation with maximum-likelihood estimation
G-computation marginalizes the conditional estimates by separating
the regression modelling from the estimation of the marginal treatment
effect for A versus C. Pass the
strategy_gcomp_ml() strategy function.
outstandR_gcomp_ml <-
outstandR(ipd_trial, ald_trial,
strategy = strategy_gcomp_ml(
formula = lin_form,
family = poisson(link = "log")))
outstandR_gcomp_ml
#> Object of class 'outstandR'
#> Model: poisson
#> Scale: log_relative_risk
#> Common treatment: C
#> Individual patient data study: AC
#> Aggregate level data study: BC
#> Confidence interval level: 0.95
#>
#> Contrasts:
#>
#> # A tibble: 3 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 AB 0.0169 0.0798 -0.537 0.571
#> 2 AC -0.877 0.0490 -1.31 -0.443
#> 3 BC -0.894 0.0308 -1.24 -0.550
#>
#> Absolute:
#>
#> # A tibble: 2 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <lgl> <lgl>
#> 1 A 0.621 0.00936 NA NA
#> 2 C 1.50 0.0717 NA NABayesian G-computation with MCMC
The difference between Bayesian G-computation and its maximum-likelihood counterpart is in the estimated distribution of the predicted outcomes. The Bayesian approach also marginalizes, integrates or standardizes over the joint posterior distribution of the conditional nuisance parameters of the outcome regression, as well as the joint covariate distribution.
Pass the strategy_gcomp_bayes() strategy function.
outstandR_gcomp_bayes <-
outstandR(ipd_trial, ald_trial,
strategy = strategy_gcomp_bayes(
formula = lin_form,
family = poisson(link = "log")))
outstandR_gcomp_bayes
#> Object of class 'outstandR'
#> Model: poisson
#> Scale: log_relative_risk
#> Common treatment: C
#> Individual patient data study: AC
#> Aggregate level data study: BC
#> Confidence interval level: 0.95
#>
#> Contrasts:
#>
#> # A tibble: 3 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 AB 0.0233 0.0851 -0.548 0.595
#> 2 AC -0.871 0.0543 -1.33 -0.414
#> 3 BC -0.894 0.0308 -1.24 -0.550
#>
#> Absolute:
#>
#> # A tibble: 2 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <lgl> <lgl>
#> 1 A 0.616 0.0102 NA NA
#> 2 C 1.48 0.0678 NA NAMultiple imputation marginalisation
Finally, the strategy function to pass to outstandR()
for multiple imputation marginalisation is
strategy_mim(),
outstandR_mim <-
outstandR(ipd_trial, ald_trial,
strategy = strategy_mim(
formula = lin_form,
family = poisson(link = "log")))#> log rate used
#>
#> log rate used
outstandR_mim
#> Object of class 'outstandR'
#> Model: poisson
#> Scale: log_relative_risk
#> Common treatment: C
#> Individual patient data study: AC
#> Aggregate level data study: BC
#> Confidence interval level: 0.95
#>
#> Contrasts:
#>
#> # A tibble: 3 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 AB 0.0237 0.0825 -0.539 0.587
#> 2 AC -0.870 0.0517 -1.32 -0.425
#> 3 BC -0.894 0.0308 -1.24 -0.550
#>
#> Absolute:
#>
#> # A tibble: 2 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <lgl> <lgl> <lgl> <lgl>
#> 1 A NA NA NA NA
#> 2 C NA NA NA NA