Introduction
This vignette will demonstrate the use of the outstandR package to fit the range of types of models to simulated binary data. Related vignettes will provide equivalent analyses for continuous and count data.
Analysis
First, let us load necessary packages.
# install.packages("outstandR",
# repos = c("https://statisticshealtheconomics.r-universe.dev", "https://cloud.r-project.org"))
#
# install.packages("simcovariates",
# repos = c("https://n8thangreen.r-universe.dev", "https://cloud.r-project.org"))
library(boot) # non-parametric bootstrap in MAIC and ML G-computation
library(copula) # simulating BC covariates from Gaussian copula
library(rstanarm) # fit outcome regression, draw outcomes in Bayesian G-computation
library(tidyr)
library(dplyr)
library(MASS)
library(outstandR)
library(simcovariates)Data
We consider binary outcomes using the log-odds ratio as the measure of effect. For example, the binary outcome may be response to treatment or the occurrence of an adverse event. For trials AC and BC, outcome for subject is simulated from a Bernoulli distribution with probabilities of success generated from logistic regression.
For the BC trial, the individual-level covariates and outcomes are aggregated to obtain summaries. The continuous covariates are summarized as means and standard deviations, which would be available to the analyst in the published study in a table of baseline characteristics in the RCT publication. The binary outcomes are summarized in an overall event table. Typically, the published study only provides aggregate information to the analyst.
The simulation input parameters are given below.
| Parameter | Description | Value |
|---|---|---|
N |
Sample size | 200 |
allocation |
Active treatment vs. placebo allocation ratio (2:1) | 2/3 |
b_trt |
Conditional effect of active treatment vs. comparator (log(0.17)) | -1.77196 |
b_X |
Conditional effect of each prognostic variable (-log(0.5)) | 0.69315 |
b_EM |
Conditional interaction effect of each effect modifier (-log(0.67)) | 0.40048 |
meanX_AC[1] |
Mean of prognostic factor in AC trial | 0.45 |
meanX_AC[2] |
Mean of prognostic factor in AC trial | 0.45 |
meanX_EM_AC[1] |
Mean of effect modifier in AC trial | 0.45 |
meanX_EM_AC[2] |
Mean of effect modifier in AC trial | 0.45 |
meanX_BC[1] |
Mean of prognostic factor in BC trial | 0.6 |
meanX_BC[2] |
Mean of prognostic factor in BC trial | 0.6 |
meanX_EM_BC[1] |
Mean of effect modifier in BC trial | 0.6 |
meanX_EM_BC[2] |
Mean of effect modifier in BC trial | 0.6 |
sdX |
Standard deviation of prognostic factors (AC and BC) | 0.4 |
sdX_EM |
Standard deviation of effect modifiers | 0.4 |
corX |
Covariate correlation coefficient | 0.2 |
b_0 |
Baseline intercept | -0.6 |
We shall use the gen_data() function available with the
simcovariates
package on GitHub.
N <- 200
allocation <- 2/3 # active treatment vs. placebo allocation ratio (2:1)
b_trt <- log(0.17) # conditional effect of active treatment vs. common comparator
b_X <- -log(0.5) # conditional effect of each prognostic variable
b_EM <- -log(0.67) # conditional interaction effect of each effect modifier
meanX_AC <- c(0.45, 0.45) # mean of normally-distributed covariate in AC trial
meanX_BC <- c(0.6, 0.6) # mean of each normally-distributed covariate in BC
meanX_EM_AC <- c(0.45, 0.45) # mean of normally-distributed EM covariate in AC trial
meanX_EM_BC <- c(0.6, 0.6) # mean of each normally-distributed EM covariate in BC
sdX <- c(0.4, 0.4) # standard deviation of each covariate (same for AC and BC)
sdX_EM <- c(0.4, 0.4) # standard deviation of each EM covariate
corX <- 0.2 # covariate correlation coefficient
b_0 <- -0.6 # baseline intercept coefficient ##TODO: fixed value
covariate_defns_ipd <- list(
PF_cont_1 = list(type = continuous(mean = meanX_AC[1], sd = sdX[1]),
role = "prognostic"),
PF_cont_2 = list(type = continuous(mean = meanX_AC[2], sd = sdX[2]),
role = "prognostic"),
EM_cont_1 = list(type = continuous(mean = meanX_EM_AC[1], sd = sdX_EM[1]),
role = "effect_modifier"),
EM_cont_2 = list(type = continuous(mean = meanX_EM_AC[2], sd = sdX_EM[2]),
role = "effect_modifier")
)
b_prognostic <- c(PF_cont_1 = b_X, PF_cont_2 = b_X)
b_effect_modifier <- c(EM_cont_1 = b_EM, EM_cont_2 = b_EM)
num_normal_covs <- length(covariate_defns_ipd)
cor_matrix <- matrix(corX, num_normal_covs, num_normal_covs)
diag(cor_matrix) <- 1
rownames(cor_matrix) <- c("PF_cont_1", "PF_cont_2", "EM_cont_1", "EM_cont_2")
colnames(cor_matrix) <- c("PF_cont_1", "PF_cont_2", "EM_cont_1", "EM_cont_2")
ipd_trial <- simcovariates::gen_data(
N = N,
b_0 = b_0,
b_trt = b_trt,
covariate_defns = covariate_defns_ipd,
b_prognostic = b_prognostic,
b_effect_modifier = b_effect_modifier,
cor_matrix = cor_matrix,
trt_assignment = list(prob_trt1 = allocation),
family = binomial("logit"))The treatment column in the return data is binary and takes values 0
and 1. We will include some extra information about treatment names. To
do this we will define the lable of the two level factor as
A for 1 and C for 0 as follows.
Similarly, to obtain the aggregate data we will simulate IPD but with
the additional summarise step. We set different mean values
meanX_BC and meanX_EM_BC but otherwise use the
same parameter values as for the
trial.
covariate_defns_ald <- list(
PF_cont_1 = list(type = continuous(mean = meanX_BC[1], sd = sdX[1]),
role = "prognostic"),
PF_cont_2 = list(type = continuous(mean = meanX_BC[2], sd = sdX[2]),
role = "prognostic"),
EM_cont_1 = list(type = continuous(mean = meanX_EM_BC[1], sd = sdX_EM[1]),
role = "effect_modifier"),
EM_cont_2 = list(type = continuous(mean = meanX_EM_BC[2], sd = sdX_EM[2]),
role = "effect_modifier")
)
BC.IPD <- simcovariates::gen_data(
N = N,
b_0 = b_0,
b_trt = b_trt,
covariate_defns = covariate_defns_ald,
b_prognostic = b_prognostic,
b_effect_modifier = b_effect_modifier,
cor_matrix = cor_matrix,
trt_assignment = list(prob_trt1 = allocation),
family = binomial("logit"))
BC.IPD$trt <- factor(BC.IPD$trt, labels = c("C", "B"))
# covariate summary statistics
# assume same between treatments
cov.X <-
BC.IPD %>%
as.data.frame() |>
dplyr::select(matches("^(PF|EM)"), trt) |>
tidyr::pivot_longer(
cols = starts_with("PF") | starts_with("EM"),
names_to = "variable",
values_to = "value") |>
group_by(variable) %>%
summarise(
mean = mean(value),
sd = sd(value)
) %>%
tidyr::pivot_longer(
cols = c("mean", "sd"),
names_to = "statistic",
values_to = "value") %>%
ungroup() |>
mutate(trt = NA)
# outcome
summary.y <-
BC.IPD |>
as.data.frame() |>
dplyr::select(y, trt) %>%
tidyr::pivot_longer(cols = "y",
names_to = "variable",
values_to = "value") %>%
group_by(variable, trt) %>%
summarise(
mean = mean(value),
sd = sd(value),
sum = sum(value)
) %>%
tidyr::pivot_longer(
cols = c("mean", "sd", "sum"),
names_to = "statistic",
values_to = "value") %>%
ungroup()
#> `summarise()` has grouped output by 'variable'. You can override using the
#> `.groups` argument.
# sample sizes
summary.N <-
BC.IPD |>
group_by(trt) |>
count(name = "N") |>
tidyr::pivot_longer(
cols = "N",
names_to = "statistic",
values_to = "value") |>
mutate(variable = NA_character_) |>
dplyr::select(variable, statistic, value, trt)
ald_trial <- rbind.data.frame(cov.X, summary.y, summary.N)This general format of the data sets are in a ‘long’ style consisting of the following.
ipd_trial: Individual patient data
-
PF_*: Patient measurements prognostic factors -
EM_*: Patient measurements effect modifiers -
trt: Treatment label (factor) -
y: Indicator of whether event was observed (two level factor)
ald_trial: Aggregate-level data
-
variable: Covariate name. In the case of treatment arm sample size this isNA -
statistic: Summary statistic name from mean, standard deviation or sum -
value: Numerical value of summary statistic -
trt: Treatment label. Because we assume a common covariate distribution between treatment arms this isNA
Our data look like the following.
head(ipd_trial)
#> id PF_cont_1 PF_cont_2 EM_cont_1 EM_cont_2 trt y true_eta
#> 1 1 0.75056436 0.95597583 0.3158969 1.19430733 C 0 0.582883524
#> 2 2 0.83246940 0.11789773 0.5094678 0.08180243 A 1 -1.476422106
#> 3 3 1.13401333 1.15036919 1.2342379 0.74771694 A 0 0.005184912
#> 4 4 0.78518298 0.83080451 -0.1625820 0.35223227 C 1 0.520117172
#> 5 5 -0.38547667 0.87070020 0.7213797 -0.03557032 A 1 -1.760974264
#> 6 6 -0.04851591 -0.03013213 0.6580067 0.05853054 A 0 -2.139514435There are 4 correlated continuous covariates generated per subject,
simulated from a multivariate normal distribution. Treatment
trt takes either new treatment A or standard of
care / status quo C. The ITC is ‘anchored’ via C, the
common treatment.
ald_trial
#> # A tibble: 16 × 4
#> variable statistic value trt
#> <chr> <chr> <dbl> <chr>
#> 1 EM_cont_1 mean 0.616 NA
#> 2 EM_cont_1 sd 0.408 NA
#> 3 EM_cont_2 mean 0.635 NA
#> 4 EM_cont_2 sd 0.354 NA
#> 5 PF_cont_1 mean 0.666 NA
#> 6 PF_cont_1 sd 0.398 NA
#> 7 PF_cont_2 mean 0.663 NA
#> 8 PF_cont_2 sd 0.404 NA
#> 9 y mean 0.597 C
#> 10 y sd 0.494 C
#> 11 y sum 43 C
#> 12 y mean 0.297 B
#> 13 y sd 0.459 B
#> 14 y sum 38 B
#> 15 NA N 72 C
#> 16 NA N 128 BIn this case, we have 4 covariate mean and standard deviation values; and the event total, average and sample size for each treatment B and C.
Regression model
The true logistic outcome model which we use to simulate the data is
is an indicator function taking value 1 if true and 0 otherwise. That
is, for treatment
the right hand side becomes
and for comparator treatments
or
there is an additional
component consisting of the effect modifier terms and the coefficient
for the treatment parameter,
(or b_trt in the R code), i.e. the log odds-ratio (LOR) for
the logit model. Finally,
is the probability of experiencing the event of interest for treatment
.
Output statistics
We will obtain the marginal treatment effect and marginal variance. The definition by which of these are calculated depends on the type of data and outcome scale. For our current example of binary data and log-odds ratio the marginal treatment effect is
and marginal variance is
where are the number of events in each arm and is the compliment of , so e.g. . Other outcome scales will be discussed below.
Model fitting in R
The outstandR package has been written to be easy to
use and essentially consists of a single function,
outstandR(). This can be used to run all of the different
types of model, which when combined with their specific parameters we
will call strategies. The first two arguments of
outstandR() are the individual patient and aggregate-level
data, respectively.
A strategy argument of outstandR takes
functions called strategy_*(), where the wildcard
* is replaced by the name of the particular method
required, e.g. strategy_maic() for MAIC. Each specific
example is provided below.
Model formula
We will take advantage of the in-built R formula object to define the models. This will allow us to easily pull out components of the object and consistently use it. Defining as effect modifiers, as prognostic variables and the treatment indicator then the formula used in this model is
Notice that this does not include the link function of interest so
appears as a linear regression. This corresponds to the following
R formula object we will pass as an argument
to the strategy function.
lin_form <- as.formula("y ~ PF_cont_1 + PF_cont_2 + trt + trt:EM_cont_1 + trt:EM_cont_2")Note that the more succinct formula
y ~ PF_cont_1 + PF_cont_2 + trt*(EM_cont_1 + EM_cont_2)would additionally include as prognostic factors so is not equivalent (but this may be what you actually want). To recover the original formula it can be modified as follows.
y ~ PF_cont_1 + PF_cont_2 + trt*(EM_cont_1 + EM_cont_2) - EM_cont_1 - EM_cont_2We note that the MAIC approach does not strictly use a regression in the same way as the other methods so should not be considered directly comparable in this sense but we have decided to use a consistent syntax across models using ‘formula’.
Matching-Adjusted Indirect Comparison (MAIC)
A single call to outstandR() is sufficient to run the
model. We pass to the strategy argument the
strategy_maic() function with arguments
formula = lin_form as defined above and
family = binomial(link = "logit") for binary data and
logistic link.
Internally, using the individual patient level data for AC
firstly we perform non-parametric bootstrap of the
maic.boot function with R = 1000 replicates.
This function fits treatment coefficient for the marginal effect for
A vs C. The returned value is an object of class
boot from the {boot} package. We then
calculate the bootstrap mean and variance in the wrapper function
maic_boot_stats.
outstandR_maic <-
outstandR(ipd_trial, ald_trial,
strategy = strategy_maic(
formula = lin_form,
family = binomial(link = "logit")))The returned object is of class outstandR.
str(outstandR_maic)
#> List of 2
#> $ contrasts:List of 3
#> ..$ means :List of 3
#> .. ..$ AB: num 0.607
#> .. ..$ AC: num -0.649
#> .. ..$ BC: num -1.26
#> ..$ variances:List of 3
#> .. ..$ AB: num 0.233
#> .. ..$ AC: num 0.137
#> .. ..$ BC: num 0.0952
#> ..$ CI :List of 3
#> .. ..$ AB: num [1:2] -0.338 1.552
#> .. ..$ AC: num [1:2] -1.3757 0.0776
#> .. ..$ BC: num [1:2] -1.861 -0.652
#> $ absolute :List of 2
#> ..$ means :List of 2
#> .. ..$ A: Named num 0.341
#> .. .. ..- attr(*, "names")= chr "mean_A"
#> .. ..$ C: Named num 0.495
#> .. .. ..- attr(*, "names")= chr "mean_C"
#> ..$ variances:List of 2
#> .. ..$ A: Named num 0.00271
#> .. .. ..- attr(*, "names")= chr "mean_A"
#> .. ..$ C: Named num 0.00518
#> .. .. ..- attr(*, "names")= chr "mean_C"
#> - attr(*, "CI")= num 0.95
#> - attr(*, "ref_trt")= chr "C"
#> - attr(*, "scale")= chr "log_odds"
#> - attr(*, "model")= chr "binomial"
#> - attr(*, "class")= chr [1:2] "outstandR" "list"We see that this is a list object with 2 parts. The first contains
statistics between each pair of treatments. These are the mean
contrasts, variances and confidence intervals (CI), respectively. The
default CI is for 95% but can be altered in outstandR with
the CI argument. The second element of the list contains
the absolute effect estimates.
A print method is available for outstandR
objects for more human-readable output
outstandR_maic
#> Object of class 'outstandR'
#> Model: binomial
#> Scale: log_odds
#> Common treatment: C
#> Individual patient data study: AC
#> Aggregate level data study: BC
#> Confidence interval level: 0.95
#>
#> Contrasts:
#>
#> # A tibble: 3 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 AB 0.607 0.233 -0.338 1.55
#> 2 AC -0.649 0.137 -1.38 0.0776
#> 3 BC -1.26 0.0952 -1.86 -0.652
#>
#> Absolute:
#>
#> # A tibble: 2 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <lgl> <lgl>
#> 1 A 0.341 0.00271 NA NA
#> 2 C 0.495 0.00518 NA NAOutcome scale
If we do not explicitly specify the outcome scale, the default is
that used to fit to the data in the regression model. As we saw, in this
case, the default is log-odds ratio corresponding to the
"logit" link function for binary data. However, we can
change this to some other scale which may be more appropriate for a
particular analysis. So far implemented in the package, the links and
their corresponding relative treatment effect scales are as follows:
| Data Type | Model | Scale | Argument |
|---|---|---|---|
| Binary | logit |
Log-odds ratio | log_odds |
| Count | log |
Log-risk ratio | log_relative_risk |
| Continuous | mean |
Mean difference | risk_difference |
The full list of possible transformed treatment effect scales will be: log-odds ratio, log-risk ratio, mean difference, risk difference, hazard ratio, hazard difference.
For binary data the marginal treatment effect and variance are
- Log-risk ratio
Treatment effect is
and variance
- Risk difference
Treatment effect is
and variance
To change the outcome scale, we can pass the scale
argument in the outstandR() function. For example, to
change the scale to risk difference, we can use the following code.
outstandR_maic_lrr <-
outstandR(ipd_trial, ald_trial,
strategy = strategy_maic(formula = lin_form,
family = binomial(link = "logit")),
scale = "log_relative_risk")
outstandR_maic_lrr
#> Object of class 'outstandR'
#> Model: binomial
#> Scale: log_relative_risk
#> Common treatment: C
#> Individual patient data study: AC
#> Aggregate level data study: BC
#> Confidence interval level: 0.95
#>
#> Contrasts:
#>
#> # A tibble: 3 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 AB 0.307 0.0793 -0.244 0.859
#> 2 AC -0.392 0.0514 -0.836 0.0527
#> 3 BC -0.699 0.0279 -1.03 -0.372
#>
#> Absolute:
#>
#> # A tibble: 2 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <lgl> <lgl>
#> 1 A 0.339 0.00258 NA NA
#> 2 C 0.501 0.00537 NA NASimulated treatment comparison (STC)
Simulated treatment comparison (STC) is the conventional outcome regression method. It involves fitting a regression model of outcome on treatment and covariates to the IPD anad then ‘plugging-in’ mean covariate values from the aggregate data study.
We can simply pass the same formula as before to the modified call
with strategy_stc().
outstandR_stc <-
outstandR(ipd_trial, ald_trial,
strategy = strategy_stc(
formula = lin_form,
family = binomial(link = "logit")))
outstandR_stc
#> Object of class 'outstandR'
#> Model: binomial
#> Scale: log_odds
#> Common treatment: C
#> Individual patient data study: AC
#> Aggregate level data study: BC
#> Confidence interval level: 0.95
#>
#> Contrasts:
#>
#> # A tibble: 3 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 AB 0.0584 NA NA NA
#> 2 AC -1.20 NA NA NA
#> 3 BC -1.26 0.0952 -1.86 -0.652
#>
#> Absolute:
#>
#> # A tibble: 2 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <lgl> <lgl>
#> 1 A 0.137 NA NA NA
#> 2 C 0.344 NA NA NAChanging the outcome scale to LRR gives
outstandR_stc_lrr <-
outstandR(ipd_trial, ald_trial,
strategy = strategy_stc(
formula = lin_form,
family = binomial(link = "logit")),
scale = "log_relative_risk")
outstandR_stc_lrr
#> Object of class 'outstandR'
#> Model: binomial
#> Scale: log_relative_risk
#> Common treatment: C
#> Individual patient data study: AC
#> Aggregate level data study: BC
#> Confidence interval level: 0.95
#>
#> Contrasts:
#>
#> # A tibble: 3 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 AB -0.224 NA NA NA
#> 2 AC -0.923 NA NA NA
#> 3 BC -0.699 0.0279 -1.03 -0.372
#>
#> Absolute:
#>
#> # A tibble: 2 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <lgl> <lgl>
#> 1 A 0.137 NA NA NA
#> 2 C 0.344 NA NA NAThe following approaches follow naturally from the simple ‘plug-in’ version of STC. We will perform G-computation firstly with a frequentist MLE and then a Bayesian approach.
Parametric G-computation with maximum-likelihood estimation
G-computation marginalizes the conditional estimates by separating the regression modelling from the estimation of the marginal treatment effect for A versus C. First, a regression model of the observed outcome on the covariates and treatment is fitted to the AC IPD:
In the context of G-computation, this regression model is often called the “Q-model.” Having fitted the Q-model, the regression coefficients are treated as nuisance parameters. The parameters are applied to the simulated covariates to predict hypothetical outcomes for each subject under both possible treatments. Namely, a pair of predicted outcomes, also called potential outcomes, under A and under C, is generated for each subject.
By plugging treatment C into the regression fit for every simulated observation, we predict the marginal outcome mean in the hypothetical scenario in which all units are under treatment C:
As performed for the previous approaches, call
outstandR() but change the strategy to
strategy_gcomp_ml(),
outstandR_gcomp_ml <-
outstandR(ipd_trial, ald_trial,
strategy = strategy_gcomp_ml(
formula = lin_form,
family = binomial(link = "logit")))
outstandR_gcomp_ml
#> Object of class 'outstandR'
#> Model: binomial
#> Scale: log_odds
#> Common treatment: C
#> Individual patient data study: AC
#> Aggregate level data study: BC
#> Confidence interval level: 0.95
#>
#> Contrasts:
#>
#> # A tibble: 3 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 AB 0.602 0.232 -0.342 1.55
#> 2 AC -0.654 0.137 -1.38 0.0714
#> 3 BC -1.26 0.0952 -1.86 -0.652
#>
#> Absolute:
#>
#> # A tibble: 2 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <lgl> <lgl>
#> 1 A 0.411 0.00291 NA NA
#> 2 C 0.570 0.00563 NA NAOnce again, let us change the outcome scale to LRR
outstandR_gcomp_ml_lrr <-
outstandR(ipd_trial, ald_trial,
strategy = strategy_gcomp_ml(
formula = lin_form,
family = binomial(link = "logit")),
scale = "log_relative_risk")
outstandR_gcomp_ml_lrr
#> Object of class 'outstandR'
#> Model: binomial
#> Scale: log_relative_risk
#> Common treatment: C
#> Individual patient data study: AC
#> Aggregate level data study: BC
#> Confidence interval level: 0.95
#>
#> Contrasts:
#>
#> # A tibble: 3 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 AB 0.377 0.0602 -0.104 0.858
#> 2 AC -0.322 0.0323 -0.674 0.0307
#> 3 BC -0.699 0.0279 -1.03 -0.372
#>
#> Absolute:
#>
#> # A tibble: 2 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <lgl> <lgl>
#> 1 A 0.409 0.00276 NA NA
#> 2 C 0.563 0.00504 NA NABayesian G-computation with MCMC
The difference between Bayesian G-computation and its maximum-likelihood counterpart is in the estimated distribution of the predicted outcomes. The Bayesian approach also marginalizes, integrates or standardizes over the joint posterior distribution of the conditional nuisance parameters of the outcome regression, as well as the joint covariate distribution.
Draw a vector of size of predicted outcomes under each set intervention from its posterior predictive distribution under the specific treatment. This is defined as where is the posterior distribution of the outcome regression coefficients , which encode the predictor-outcome relationships observed in the AC trial IPD. This is given by:
In practice, the integrals above can be approximated numerically, using full Bayesian estimation via Markov chain Monte Carlo (MCMC) sampling.
The average, variance and interval estimates of the marginal treatment effect can be derived empirically from draws of the posterior density.
The strategy function to plug-in to the outstandR() call
for this approach is strategy_gcomp_bayes(),
outstandR_gcomp_bayes <-
outstandR(ipd_trial, ald_trial,
strategy = strategy_gcomp_bayes(
formula = lin_form,
family = binomial(link = "logit")))
outstandR_gcomp_bayes
#> Object of class 'outstandR'
#> Model: binomial
#> Scale: log_odds
#> Common treatment: C
#> Individual patient data study: AC
#> Aggregate level data study: BC
#> Confidence interval level: 0.95
#>
#> Contrasts:
#>
#> # A tibble: 3 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 AB 0.612 0.228 -0.325 1.55
#> 2 AC -0.644 0.133 -1.36 0.0713
#> 3 BC -1.26 0.0952 -1.86 -0.652
#>
#> Absolute:
#>
#> # A tibble: 2 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <lgl> <lgl>
#> 1 A 0.411 0.00288 NA NA
#> 2 C 0.568 0.00521 NA NAAs before, we can change the outcome scale to LRR.
outstandR_gcomp_bayes_lrr <-
outstandR(ipd_trial, ald_trial,
strategy = strategy_gcomp_bayes(
formula = lin_form,
family = binomial(link = "logit")),
scale = "log_relative_risk")
outstandR_gcomp_bayes_lrr
#> Object of class 'outstandR'
#> Model: binomial
#> Scale: log_relative_risk
#> Common treatment: C
#> Individual patient data study: AC
#> Aggregate level data study: BC
#> Confidence interval level: 0.95
#>
#> Contrasts:
#>
#> # A tibble: 3 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 AB 0.375 0.0607 -0.108 0.858
#> 2 AC -0.324 0.0328 -0.679 0.0308
#> 3 BC -0.699 0.0279 -1.03 -0.372
#>
#> Absolute:
#>
#> # A tibble: 2 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <lgl> <lgl>
#> 1 A 0.411 0.00288 NA NA
#> 2 C 0.568 0.00521 NA NAMultiple imputation marginalisation
The final method is to obtain the marginalized treatment effect for aggregate level data study, obtained by integrating over the covariate distribution from the aggregate level data study
The aggregate level data likelihood is
The multiple imputation marginalisation strategy function is
strategy_mim(),
outstandR_mim <-
outstandR(ipd_trial, ald_trial,
strategy = strategy_mim(
formula = lin_form,
family = binomial(link = "logit")))
outstandR_mim
#> Object of class 'outstandR'
#> Model: binomial
#> Scale: log_odds
#> Common treatment: C
#> Individual patient data study: AC
#> Aggregate level data study: BC
#> Confidence interval level: 0.95
#>
#> Contrasts:
#>
#> # A tibble: 3 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 AB 0.612 0.216 -0.299 1.52
#> 2 AC -0.644 0.121 -1.33 0.0386
#> 3 BC -1.26 0.0952 -1.86 -0.652
#>
#> Absolute:
#>
#> # A tibble: 2 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <lgl> <lgl> <lgl> <lgl>
#> 1 A NA NA NA NA
#> 2 C NA NA NA NAChange the outcome scale again for LRR,
outstandR_mim_lrr <-
outstandR(ipd_trial, ald_trial,
strategy = strategy_mim(
formula = lin_form,
family = binomial(link = "logit")),
scale = "log_relative_risk")
outstandR_mim_lrr
#> Object of class 'outstandR'
#> Model: binomial
#> Scale: log_relative_risk
#> Common treatment: C
#> Individual patient data study: AC
#> Aggregate level data study: BC
#> Confidence interval level: 0.95
#>
#> Contrasts:
#>
#> # A tibble: 3 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 AB 0.375 0.0516 -0.0704 0.820
#> 2 AC -0.324 0.0237 -0.626 -0.0227
#> 3 BC -0.699 0.0279 -1.03 -0.372
#>
#> Absolute:
#>
#> # A tibble: 2 × 5
#> Treatments Estimate Std.Error lower.0.95 upper.0.95
#> <chr> <lgl> <lgl> <lgl> <lgl>
#> 1 A NA NA NA NA
#> 2 C NA NA NA NAModel comparison
effect in population
The true effect on the log OR scale in the (aggregate trial data) population is . Calculated by
mean_X1 <- ald_trial$value[ald_trial$statistic == "mean" & ald_trial$variable == "EM_cont_1"]
mean_X2 <- ald_trial$value[ald_trial$statistic == "mean" & ald_trial$variable == "EM_cont_1"]
d_AC_true <- b_trt + b_EM * (mean_X1 + mean_X2)The naive approach is to just convert directly from one population to another, ignoring the imbalance in effect modifiers.
d_AC_naive <-
ipd_trial |>
dplyr::group_by(trt) |>
dplyr::summarise(y_sum = sum(y), y_bar = mean(y), n = n()) |>
tidyr::pivot_wider(names_from = trt,
values_from = c(y_sum, y_bar, n)) |>
dplyr::mutate(d_AC =
log(y_bar_A/(1-y_bar_A)) - log(y_bar_C/(1-y_bar_C)),
var_AC =
1/(n_A-y_sum_A) + 1/y_sum_A + 1/(n_C-y_sum_C) + 1/y_sum_C)effect in population
This is the indirect effect. The true effect in the population is .
Following the simulation study in Remiro et al (2020) these cancel out and the true effect is zero.
The naive comparison calculating effect in the population is
# reshape to make extraction easier
ald_out <- ald_trial |>
dplyr::filter(variable == "y" | is.na(variable)) |>
mutate(stat_trt = paste0(statistic, "_", trt)) |>
dplyr::select(stat_trt, value) |>
pivot_wider(names_from = stat_trt, values_from = value)
d_BC <-
with(ald_out, log(mean_B/(1-mean_B)) - log(mean_C/(1-mean_C)))
d_AB_naive <- d_AC_naive$d_AC - d_BC
var.d.BC <- with(ald_out, 1/sum_B + 1/(N_B - sum_B) + 1/sum_C + 1/(N_C - sum_C))
var.d.AB.naive <- d_AC_naive$var_AC + var.d.BCOf course, the contrast is calculated directly.
Results
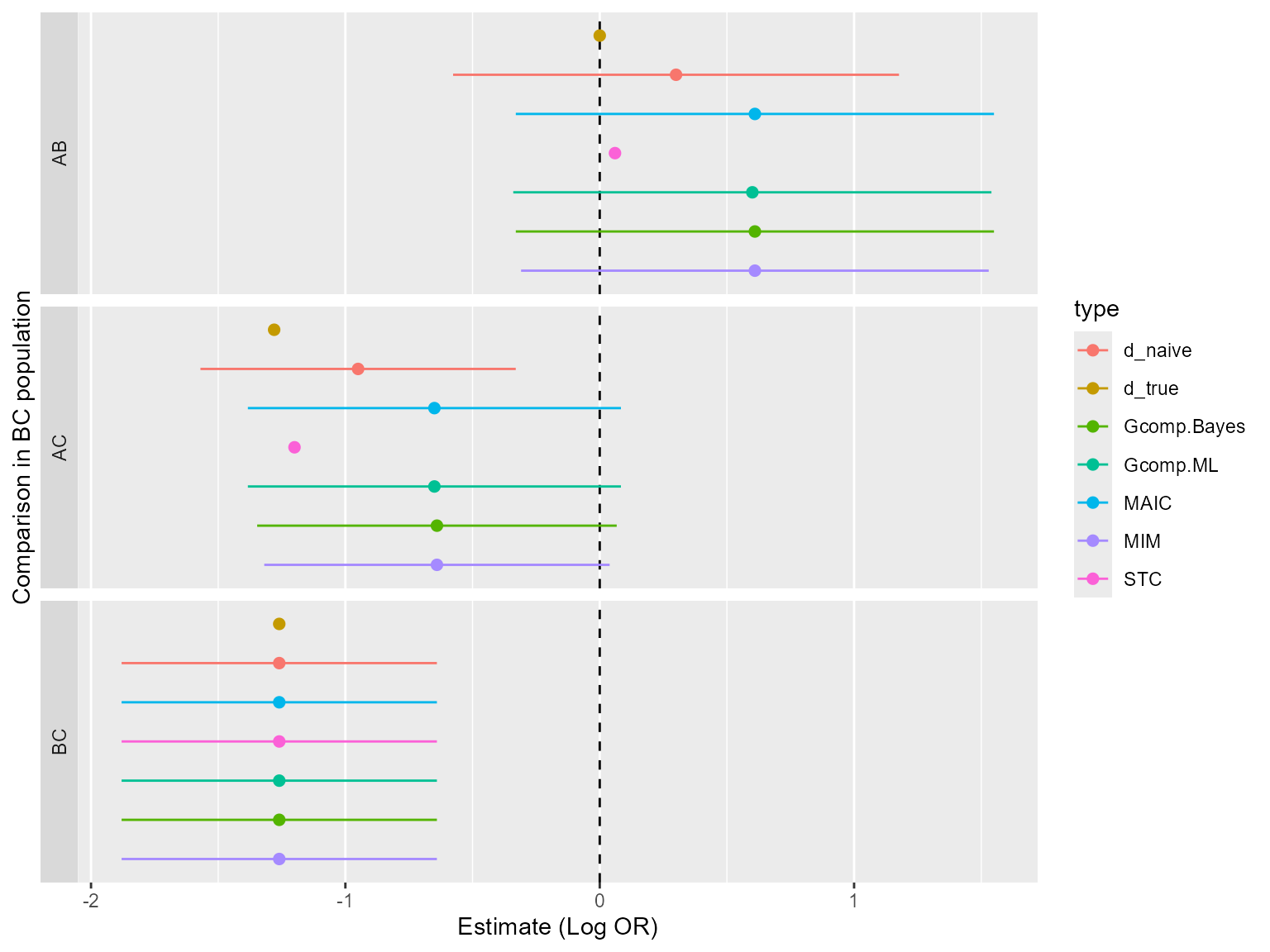
We now combine all outputs and present in plots and tables. For a log-odds ratio a table of all contrasts and methods in the population is given below.
| d_true | d_naive | MAIC | STC | Gcomp.ML | Gcomp.Bayes | MIM | |
|---|---|---|---|---|---|---|---|
| AB | 0.00 | 0.30 | 0.61 | 0.06 | 0.60 | 0.61 | 0.61 |
| AC | -1.28 | -0.95 | -0.65 | -1.20 | -0.65 | -0.64 | -0.64 |
| BC | -1.26 | -1.26 | -1.26 | -1.26 | -1.26 | -1.26 | -1.26 |
We can see that the different corresponds reasonably well with one another.
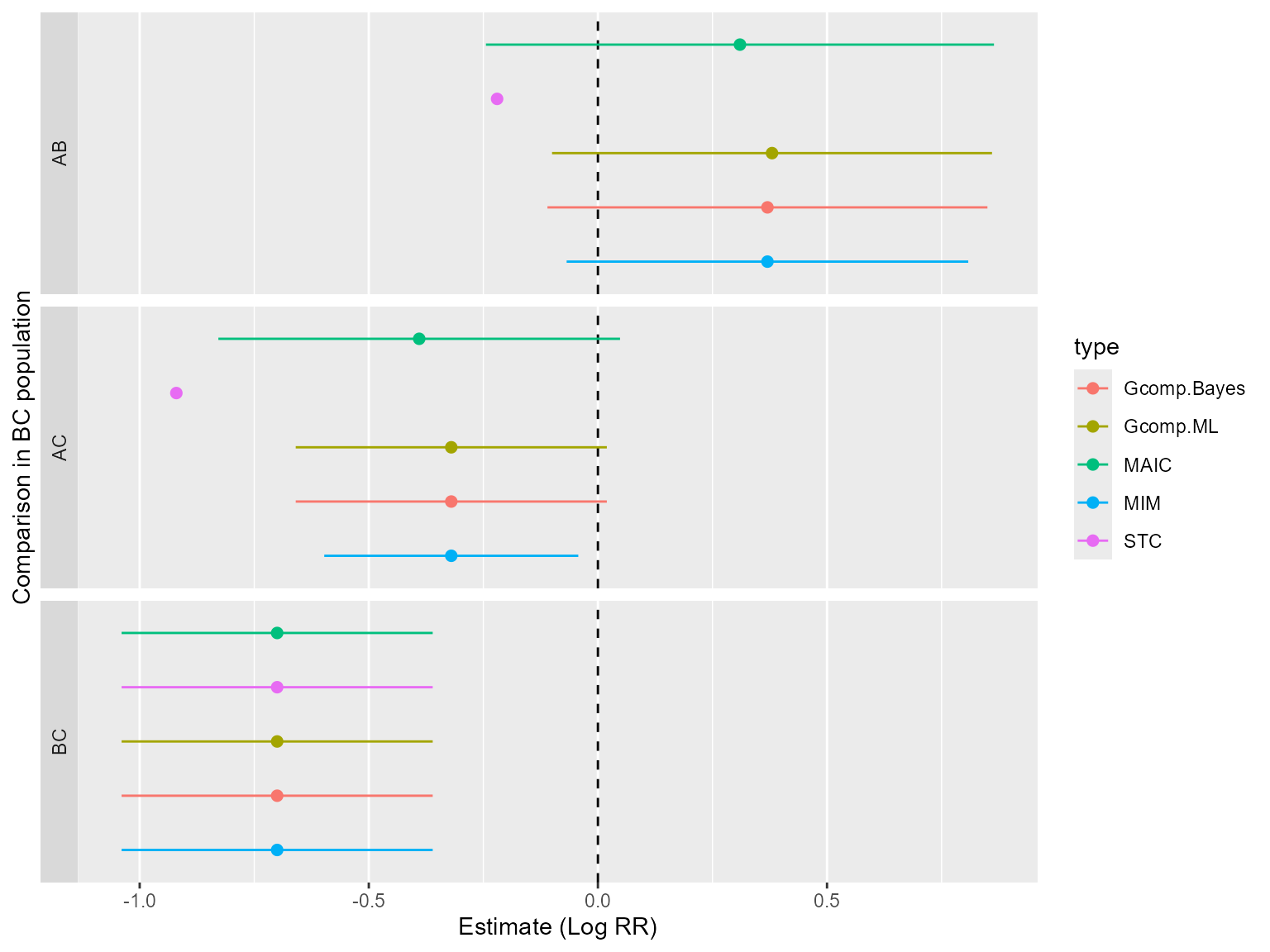
Next, let us look at the results on the log relative risk scale. First, the true values are calculated as
so that the summary table is
| MAIC | STC | Gcomp.ML | Gcomp.Bayes | MIM | |
|---|---|---|---|---|---|
| AB | 0.31 | -0.22 | 0.38 | 0.37 | 0.37 |
| AC | -0.39 | -0.92 | -0.32 | -0.32 | -0.32 |
| BC | -0.70 | -0.70 | -0.70 | -0.70 | -0.70 |