INLA piece-wise exponential and external knowledge
First load the package data and take a look.
data("TA174_FCR", package = "blendR")
head(dat_FCR)
#> # A tibble: 6 × 5
#> patid treat death death_t death_ty
#> <int> <int> <int> <dbl> <dbl>
#> 1 1 1 0 32 2.67
#> 2 2 1 0 30.6 2.55
#> 3 3 1 0 28 2.33
#> 4 8 1 0 30 2.5
#> 5 10 1 1 0.458 0.0382
#> 6 11 1 1 1.57 0.131Next, fit the observed data with a piece-wise exponential model using INLA.
## observed estimate
obs_Surv <- fit_inla_pw(data = dat_FCR,
cutpoints = seq(0, 180, by = 5))For the external model first we create some synthetic data consistent with user-defined constraints as follows.
## external estimate
data_sim <- ext_surv_sim(t_info = 144,
S_info = 0.05,
T_max = 180)Then fit a gompertz model to this.
ext_Surv <- fit.models(formula = Surv(time, event) ~ 1,
data = data_sim,
distr = "gompertz",
method = "hmc",
priors = list(gom = list(a_alpha = 0.1,
b_alpha = 0.1)))
#>
#> SAMPLING FOR MODEL 'Gompertz' NOW (CHAIN 1).
#> Chain 1:
#> Chain 1: Gradient evaluation took 5.1e-05 seconds
#> Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.51 seconds.
#> Chain 1: Adjust your expectations accordingly!
#> Chain 1:
#> Chain 1:
#> Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
#> Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
#> Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
#> Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
#> Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
#> Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
#> Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
#> Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
#> Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
#> Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
#> Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
#> Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
#> Chain 1:
#> Chain 1: Elapsed Time: 0.581 seconds (Warm-up)
#> Chain 1: 0.305 seconds (Sampling)
#> Chain 1: 0.886 seconds (Total)
#> Chain 1:
#>
#> SAMPLING FOR MODEL 'Gompertz' NOW (CHAIN 2).
#> Chain 2: Rejecting initial value:
#> Chain 2: Log probability evaluates to log(0), i.e. negative infinity.
#> Chain 2: Stan can't start sampling from this initial value.
#> Chain 2: Rejecting initial value:
#> Chain 2: Log probability evaluates to log(0), i.e. negative infinity.
#> Chain 2: Stan can't start sampling from this initial value.
#> Chain 2:
#> Chain 2: Gradient evaluation took 2.8e-05 seconds
#> Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.28 seconds.
#> Chain 2: Adjust your expectations accordingly!
#> Chain 2:
#> Chain 2:
#> Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
#> Chain 2: Iteration: 200 / 2000 [ 10%] (Warmup)
#> Chain 2: Iteration: 400 / 2000 [ 20%] (Warmup)
#> Chain 2: Iteration: 600 / 2000 [ 30%] (Warmup)
#> Chain 2: Iteration: 800 / 2000 [ 40%] (Warmup)
#> Chain 2: Iteration: 1000 / 2000 [ 50%] (Warmup)
#> Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
#> Chain 2: Iteration: 1200 / 2000 [ 60%] (Sampling)
#> Chain 2: Iteration: 1400 / 2000 [ 70%] (Sampling)
#> Chain 2: Iteration: 1600 / 2000 [ 80%] (Sampling)
#> Chain 2: Iteration: 1800 / 2000 [ 90%] (Sampling)
#> Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
#> Chain 2:
#> Chain 2: Elapsed Time: 0.98 seconds (Warm-up)
#> Chain 2: 0.245 seconds (Sampling)
#> Chain 2: 1.225 seconds (Total)
#> Chain 2:Lastly, we can run the blending step.
blend_interv <- list(min = 48, max = 150)
beta_params <- list(alpha = 3, beta = 3)
ble_Surv <- blendsurv(obs_Surv, ext_Surv, blend_interv, beta_params)
#> 'as(<dgCMatrix>, "dgTMatrix")' is deprecated.
#> Use 'as(., "TsparseMatrix")' instead.
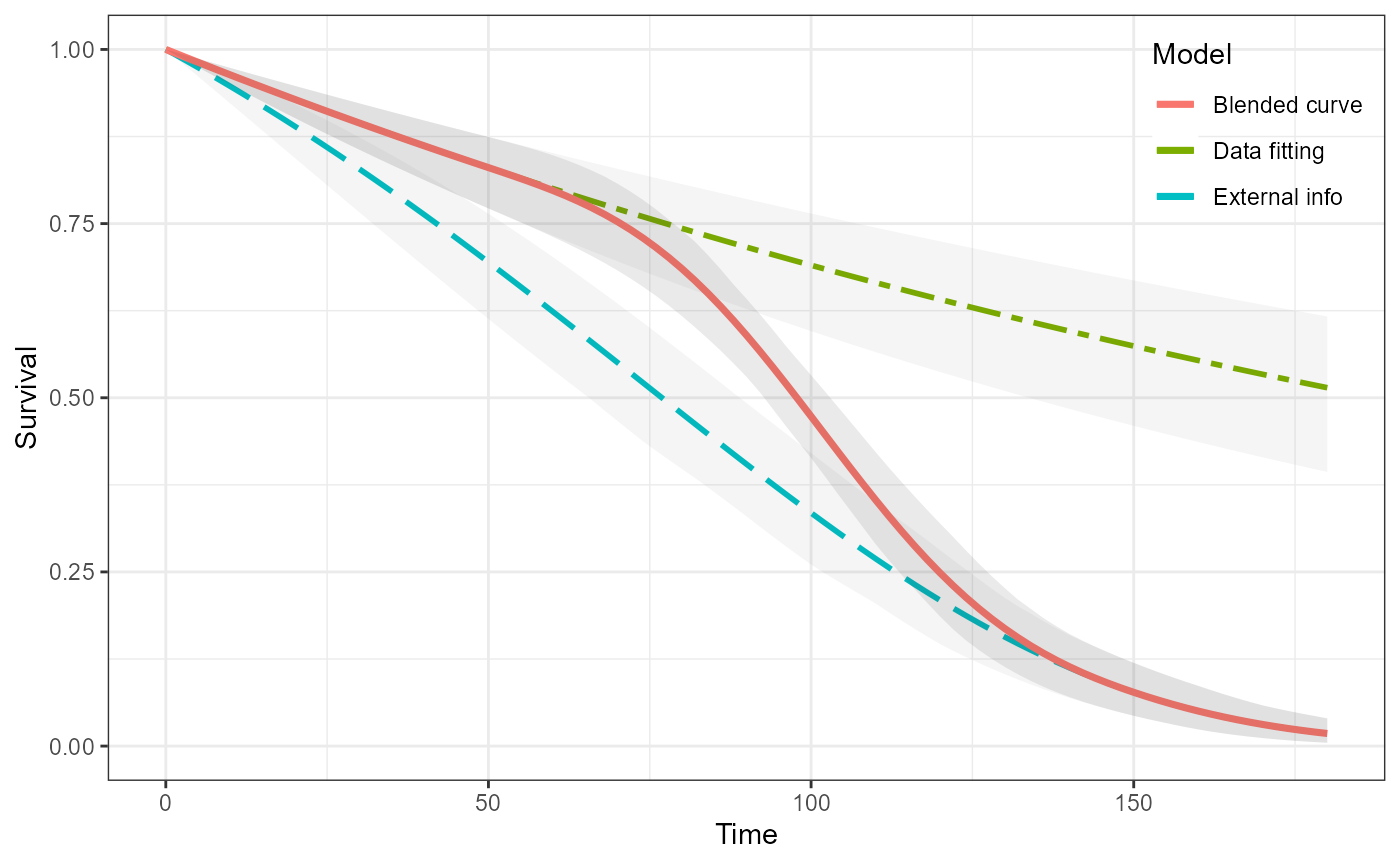
#> See help("Deprecated") and help("Matrix-deprecated").We can visualise all of the curves.
plot(ble_Surv)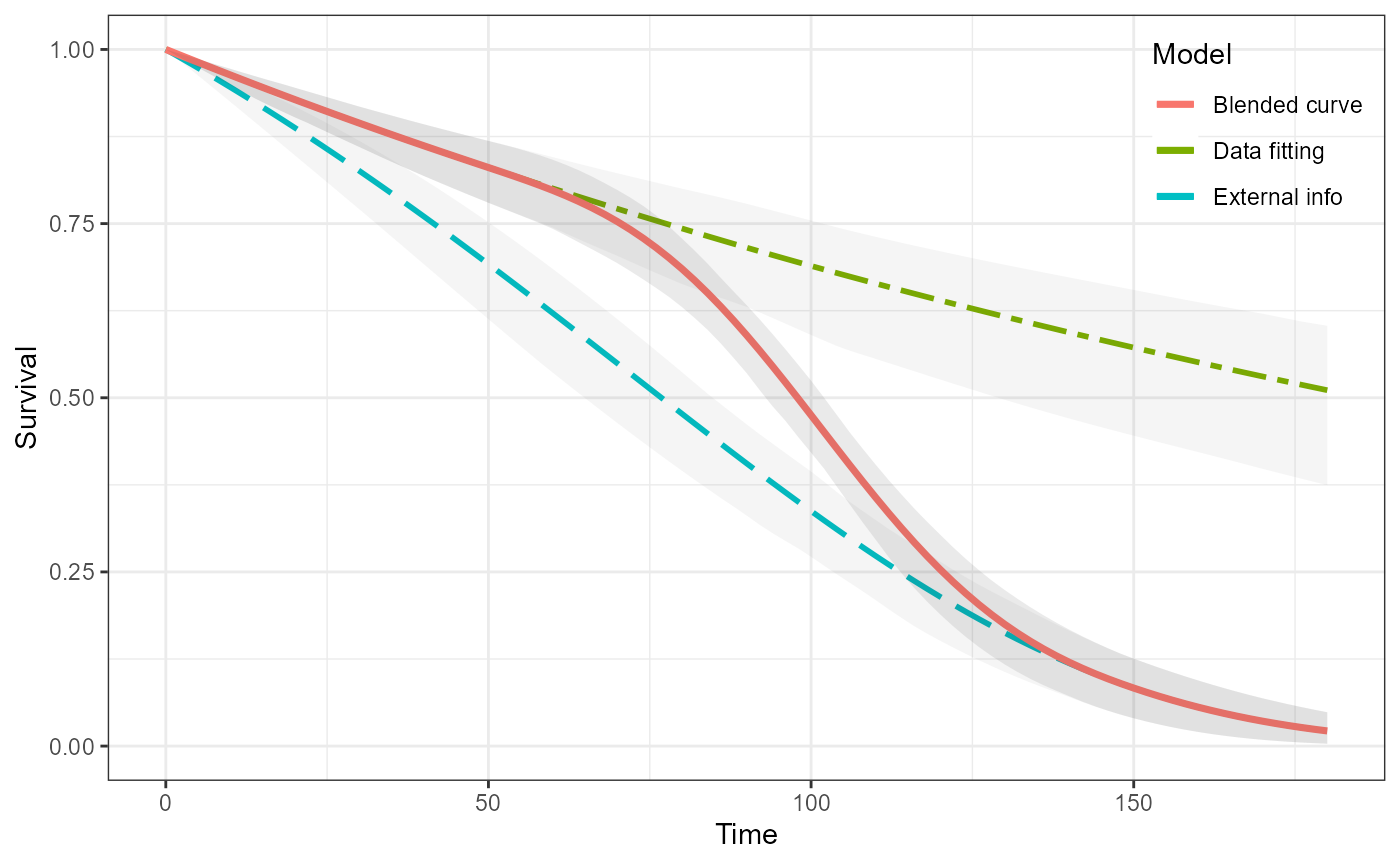
Two HMC survHE fits
In the same way as above we fit two survival curves and then blend them together. In this case we use two exponential curves using the survHE package to fit them.
obs_Surv2 <- fit.models(formula = Surv(death_t, death) ~ 1,
data = dat_FCR,
distr = "exponential",
method = "hmc")
#>
#> SAMPLING FOR MODEL 'Exponential' NOW (CHAIN 1).
#> Chain 1:
#> Chain 1: Gradient evaluation took 6.7e-05 seconds
#> Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.67 seconds.
#> Chain 1: Adjust your expectations accordingly!
#> Chain 1:
#> Chain 1:
#> Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
#> Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
#> Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
#> Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
#> Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
#> Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
#> Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
#> Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
#> Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
#> Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
#> Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
#> Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
#> Chain 1:
#> Chain 1: Elapsed Time: 0.417 seconds (Warm-up)
#> Chain 1: 0.289 seconds (Sampling)
#> Chain 1: 0.706 seconds (Total)
#> Chain 1:
#>
#> SAMPLING FOR MODEL 'Exponential' NOW (CHAIN 2).
#> Chain 2:
#> Chain 2: Gradient evaluation took 6.4e-05 seconds
#> Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.64 seconds.
#> Chain 2: Adjust your expectations accordingly!
#> Chain 2:
#> Chain 2:
#> Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
#> Chain 2: Iteration: 200 / 2000 [ 10%] (Warmup)
#> Chain 2: Iteration: 400 / 2000 [ 20%] (Warmup)
#> Chain 2: Iteration: 600 / 2000 [ 30%] (Warmup)
#> Chain 2: Iteration: 800 / 2000 [ 40%] (Warmup)
#> Chain 2: Iteration: 1000 / 2000 [ 50%] (Warmup)
#> Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
#> Chain 2: Iteration: 1200 / 2000 [ 60%] (Sampling)
#> Chain 2: Iteration: 1400 / 2000 [ 70%] (Sampling)
#> Chain 2: Iteration: 1600 / 2000 [ 80%] (Sampling)
#> Chain 2: Iteration: 1800 / 2000 [ 90%] (Sampling)
#> Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
#> Chain 2:
#> Chain 2: Elapsed Time: 0.434 seconds (Warm-up)
#> Chain 2: 0.266 seconds (Sampling)
#> Chain 2: 0.7 seconds (Total)
#> Chain 2:
ext_Surv2 <- fit.models(formula = Surv(time, event) ~ 1,
data = data_sim,
distr = "exponential",
method = "hmc")
#>
#> SAMPLING FOR MODEL 'Exponential' NOW (CHAIN 1).
#> Chain 1:
#> Chain 1: Gradient evaluation took 2.2e-05 seconds
#> Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.22 seconds.
#> Chain 1: Adjust your expectations accordingly!
#> Chain 1:
#> Chain 1:
#> Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
#> Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
#> Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
#> Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
#> Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
#> Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
#> Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
#> Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
#> Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
#> Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
#> Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
#> Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
#> Chain 1:
#> Chain 1: Elapsed Time: 0.173 seconds (Warm-up)
#> Chain 1: 0.071 seconds (Sampling)
#> Chain 1: 0.244 seconds (Total)
#> Chain 1:
#>
#> SAMPLING FOR MODEL 'Exponential' NOW (CHAIN 2).
#> Chain 2:
#> Chain 2: Gradient evaluation took 2.5e-05 seconds
#> Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.25 seconds.
#> Chain 2: Adjust your expectations accordingly!
#> Chain 2:
#> Chain 2:
#> Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
#> Chain 2: Iteration: 200 / 2000 [ 10%] (Warmup)
#> Chain 2: Iteration: 400 / 2000 [ 20%] (Warmup)
#> Chain 2: Iteration: 600 / 2000 [ 30%] (Warmup)
#> Chain 2: Iteration: 800 / 2000 [ 40%] (Warmup)
#> Chain 2: Iteration: 1000 / 2000 [ 50%] (Warmup)
#> Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
#> Chain 2: Iteration: 1200 / 2000 [ 60%] (Sampling)
#> Chain 2: Iteration: 1400 / 2000 [ 70%] (Sampling)
#> Chain 2: Iteration: 1600 / 2000 [ 80%] (Sampling)
#> Chain 2: Iteration: 1800 / 2000 [ 90%] (Sampling)
#> Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
#> Chain 2:
#> Chain 2: Elapsed Time: 0.149 seconds (Warm-up)
#> Chain 2: 0.071 seconds (Sampling)
#> Chain 2: 0.22 seconds (Total)
#> Chain 2:
ble_Surv2 <- blendsurv(obs_Surv2, ext_Surv2, blend_interv, beta_params)
# kaplan-meier
km <- survfit(Surv(death_t, death) ~ 1, data = dat_FCR)
plot(ble_Surv2) +
geom_line(aes(km$time, km$surv, colour = "Kaplan-Meier"),
size = 1.25, linetype = "dashed")
#> Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
#> ℹ Please use `linewidth` instead.
#> This warning is displayed once every 8 hours.
#> Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
#> generated.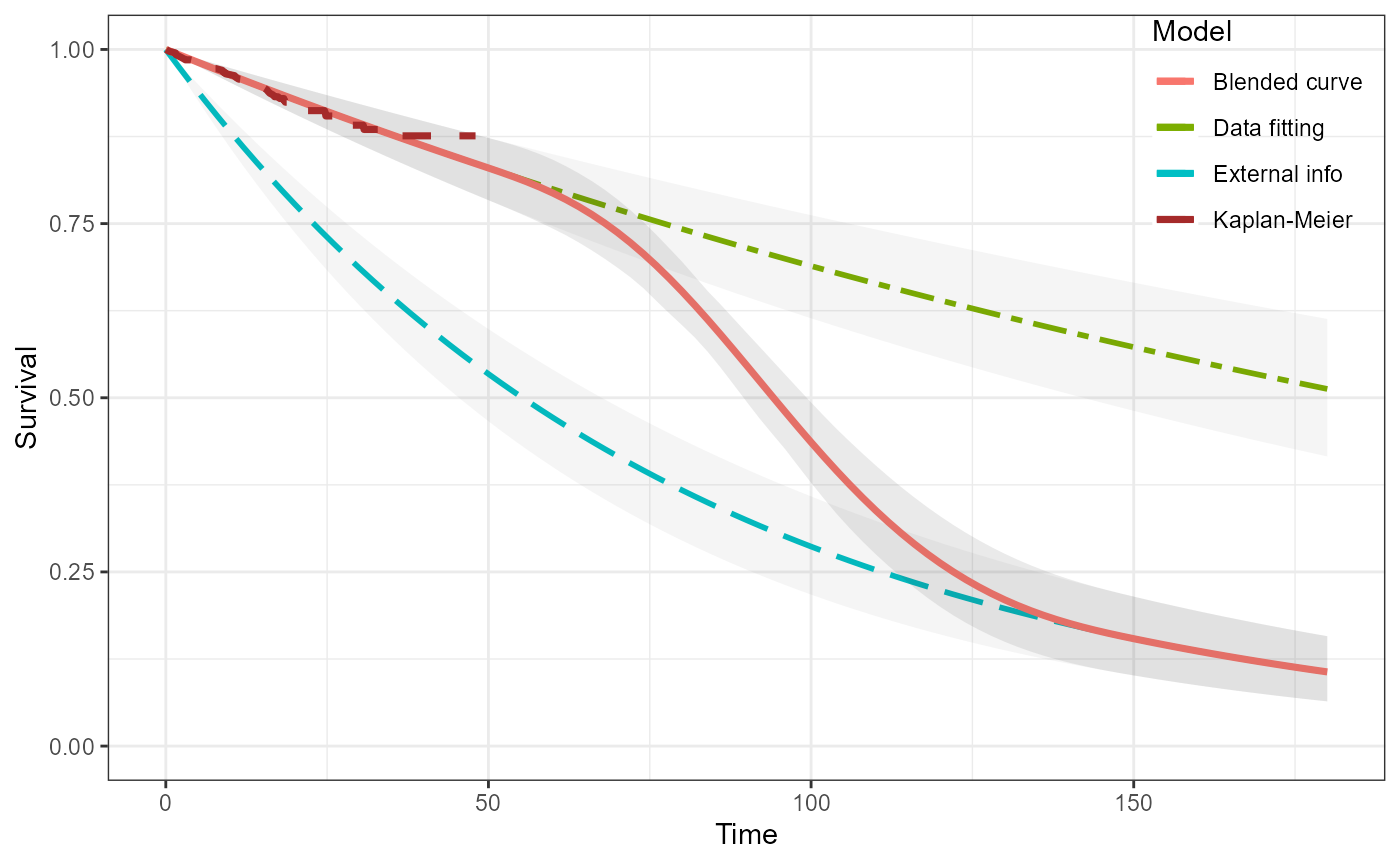
flexsurv frequentist background fit
The last example is for an HMC and frequentist survival model using the flexsurv package directly.
obs_Surv3 <- fit.models(formula = Surv(death_t, death) ~ 1,
data = dat_FCR,
distr = "exponential",
method = "hmc")
#>
#> SAMPLING FOR MODEL 'Exponential' NOW (CHAIN 1).
#> Chain 1:
#> Chain 1: Gradient evaluation took 5.6e-05 seconds
#> Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.56 seconds.
#> Chain 1: Adjust your expectations accordingly!
#> Chain 1:
#> Chain 1:
#> Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
#> Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
#> Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
#> Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
#> Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
#> Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
#> Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
#> Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
#> Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
#> Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
#> Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
#> Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
#> Chain 1:
#> Chain 1: Elapsed Time: 0.405 seconds (Warm-up)
#> Chain 1: 0.257 seconds (Sampling)
#> Chain 1: 0.662 seconds (Total)
#> Chain 1:
#>
#> SAMPLING FOR MODEL 'Exponential' NOW (CHAIN 2).
#> Chain 2:
#> Chain 2: Gradient evaluation took 5.2e-05 seconds
#> Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.52 seconds.
#> Chain 2: Adjust your expectations accordingly!
#> Chain 2:
#> Chain 2:
#> Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
#> Chain 2: Iteration: 200 / 2000 [ 10%] (Warmup)
#> Chain 2: Iteration: 400 / 2000 [ 20%] (Warmup)
#> Chain 2: Iteration: 600 / 2000 [ 30%] (Warmup)
#> Chain 2: Iteration: 800 / 2000 [ 40%] (Warmup)
#> Chain 2: Iteration: 1000 / 2000 [ 50%] (Warmup)
#> Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
#> Chain 2: Iteration: 1200 / 2000 [ 60%] (Sampling)
#> Chain 2: Iteration: 1400 / 2000 [ 70%] (Sampling)
#> Chain 2: Iteration: 1600 / 2000 [ 80%] (Sampling)
#> Chain 2: Iteration: 1800 / 2000 [ 90%] (Sampling)
#> Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
#> Chain 2:
#> Chain 2: Elapsed Time: 0.378 seconds (Warm-up)
#> Chain 2: 0.235 seconds (Sampling)
#> Chain 2: 0.613 seconds (Total)
#> Chain 2:
ext_Surv3 <- flexsurv::flexsurvreg(formula = Surv(time, event) ~ 1,
data = data_sim,
dist = "gompertz")
ble_Surv3 <- blendsurv(obs_Surv3, ext_Surv3, blend_interv, beta_params)
plot(ble_Surv3)